CS188 Project
08 Jun 2017 | machine-learning nlpLiterature Review
A literature review of some of the tools we used can be found here
Summary
Using a novel combination of LDA and Document Vectors, our project aims to assist doctor’s in creating reports by providing real-time suggestions of information doctors may have missed. Additionally, our robust machine learning model can be used to categorize and find similar documents.
Methods
Our dataset consisted of roughly 400 clinical documents each including a few pages of notes for an MRI imaging session. Using a Python script, these documents were first parsed using regular expressions and then inserted into a Mongo database for easier manipulation. We trained on 336 of these documents and set aside 50 documents as a test set. Each document was tokenized, lowercased, and stripped of all punctuation and useless general words like conjunctions, pronouns, and articles. Using our set of training documents, we created a corpus and a bag-of-words dictionary.
With our new corpus, we trained document vectors for each document[2]. To generate topics for the data, we used Latent Dirichlet Analysis (LDA) [1] to automatically discover topical themes from text documents. Preprocessing the input text is an essential part of LDA. Like most LDA topic models, we relied on a bag-of-word model for each document which reflected how often words appeared throughout a document[5]. To detect compelling topics, we discarded very common words which did not differentiate between topics and very rare words as well [4].
We also needed a phrase extractor to suggest meaningful, specific phrases well representing the document’s topic. To develop this rich vocabulary, we looked to the Python library Natrual Language ToolKit (NLTK) with its collocations class to detect bigrams based on points of mutual information(pmi) and gensim offering both bigrams and trigrams. However, these models failed to produce compelling phrases. Upon analyzing our data, we found that medical documents contained long descriptive phrases like “right basal ganglia” or “minimal paranasal sinus mucosal thickening” that were too rare to be considered a phrase yet contained words like “right” and “minimal” that possess little meaning alone. Resolving this shortcoming, we employed NLTK’s WordNet to detect parts of speech and extract noun phrases. These noun phrases reflected these long medical phrases and encapsulated their complexity.
Model/Evaluation
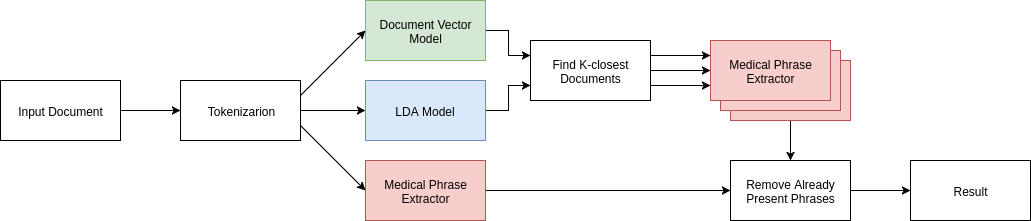
The goal of our project is to suggest phrases a doctor may have missed. So, to test our effectiveness, we set aside a test set of 50 documents from our training data. From these 50 documents, we removed several key medical phrases and noted which removed phrases our model successfully suggested.
First, we remove 10 key medical phrases from each document. Next, we pass each document to our document vector and LDA models, and get back the k most frequent documents that are within a distance threshold. We tested different k values and different distance thresholds for both the LDA model and our document vector model. We then keep a count of each medical term that is extracted from all of these documents. The top 10 most frequent medical terms are then taken as our result. We take the average number of phrases that are recovered by our method as a metric of our model’s success.
We wanted to know if our combination of document and LDA topic vectors would recover more phrases than a pipeline that only used document vectors or LDA topic vectors, so we ran our testing methodology thrice, one just using the LDA topic vectors to find similar documents, once just using the document vector to find similar documents, and once using the two in tandem.
Results
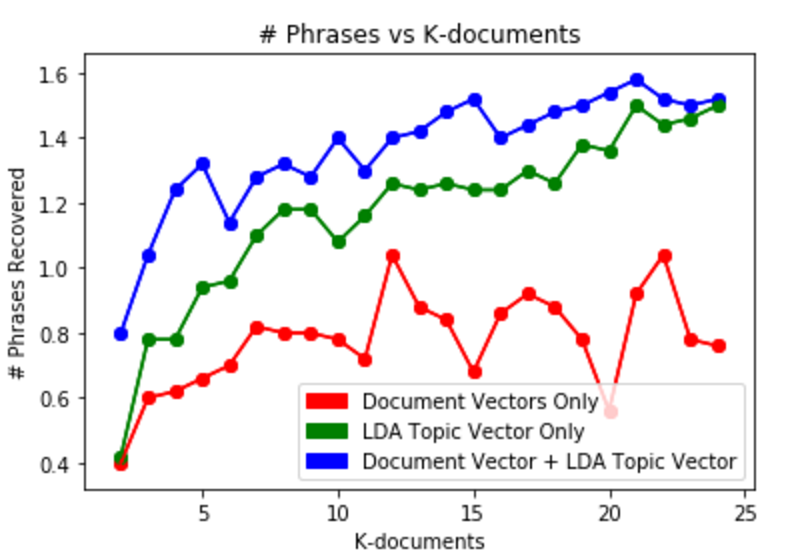
We found that on average, our model was able to recover 1.6 phrases from each medical document. Our model, which combined both document vectors and topic vectors, worked better than either approach individually. We recover nearly twice the number of medical terms from the document vector only approach and perform significantly better than the topic vector only approach when restricted to looking at a few documents.
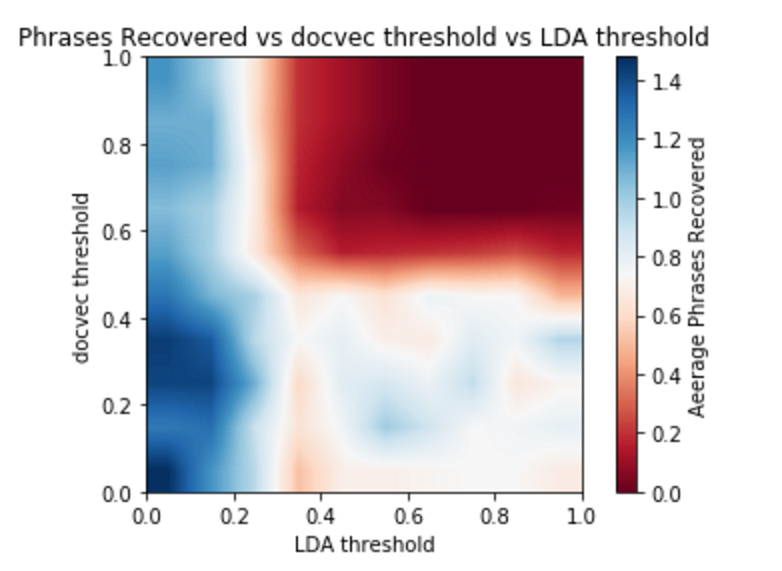
Interestingly, increasing the threshold does not increase the number of terms recovered. We feel like this is likely due to the restrictions on our training data. With a larger corpus, we would be able to get a better sense of the relationship of these thresholds. In general, we are unable to recover many, if any terms when our distance thresholds are too high. This is likely because there are simply not enough documents higher than the threshold.
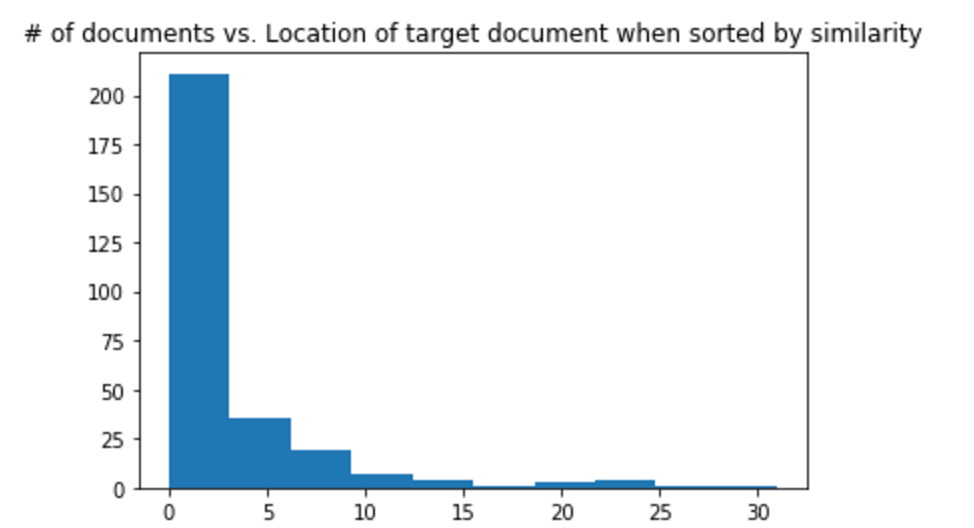
To ensure our document vectors were properly trained, we took each document in the training corpus, and found the most similar document to itself. Ideally, if our document vector model is working well, each document should be most similar to itself. We found that around 70% of our training corpus was most similar to itself. We also manually looked at the most similar document for some of the test documents in our testing set to ensure that we were not overfitting the model.
We also manually reviewed some of the phrases that our pipeline was able to recover.
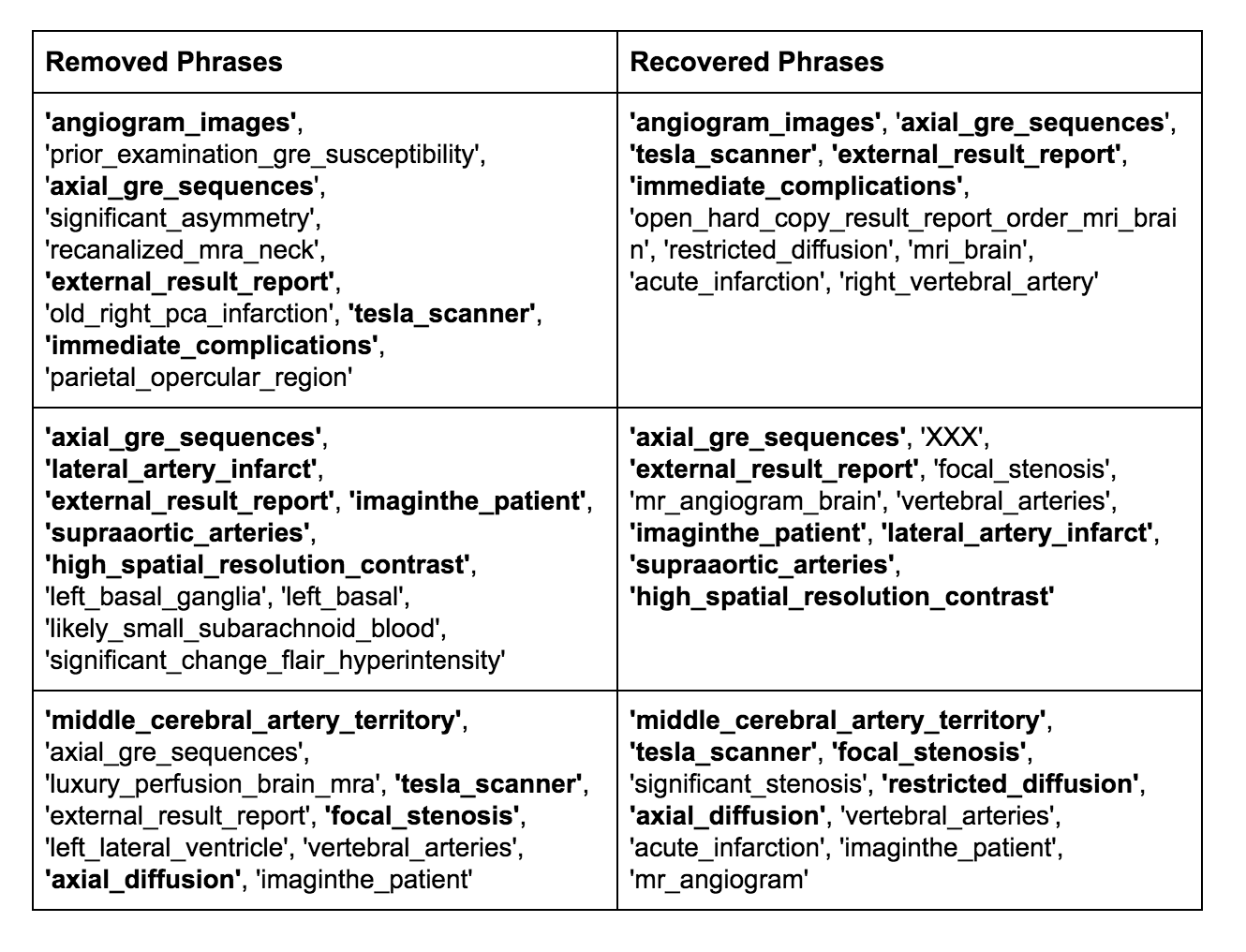
We found that while some of the phrases we recovered were just white noise, we were able to recover some significant medical information, such as ‘lateral_artery_infarct’ and ‘middle_cerebral_artery_territory’.
Conclusion
Our original goal was to try and mimic the approach used in Hoo-Chang Shin’s paper [3], but with MRI data instead of chest X-Ray data. With the help of our advisor Irma Ravkic, we were given UCLA MRI scans with detailed document notes. As in the paper, we began by building a topical model to categorize documents to later train a CNN on these labels. Unfortunately, due to a bookkeeping error, the documents we were analyzing couldn’t be matched to their corresponding images.
Nevertheless, we were able to leverage our rich textual analysis to find information that doctors may have missed. While our model currently offers simple phrase detection, we imagine that our rich analysis of medical documents offers a multitude of potential applications. Our system could easily be modified to one day diagnosis patients based merely on documents of doctor observations.
All in all, we were pleased with the results we achieved. One of the largest limitations we had was the relatively small number of documents for a relatively small number of topics. More data would produce a more accurate model applicable across the medical field.
One disadvantage of our pipeline was the manual tuning of the number of topics in our LDA model. Future implementation could use Hierarchical Dirichlet Process Latent Dirichlet Allocation (HDP-LDA) to remove this parameter and allow a more adaptive model. Another limitation of our model was our relatively rudimentary implementation of a noun phrase extraction. A more professional model could potentially improve our results, as it seemed that some our noun phrases were not strictly medical terms.
References
- Blei, D. M.; Ng, A. Y.; and Jordan, M. I: “Latent Dirichlet Allocation”. 2003. Journal of Machine Learning research 3:993–1022.
- “Doc2Vec Tutorial on the Lee Dataset.” GitHub. RaRe-Technologies, 30 Mar. 2016. Web. 05 July 2017. [https://github.com/RaRe-Technologies/gensim/blob/develop/docs/notebooks/doc2vec-lee.ipynb]
- Hoo-Chang Shin, Kirk Roberts, Le Lu, Dina Demner-Fushman, Jianhua Yao: “Learning to Read Chest X-Rays: Recurrent Neural Cascade Model for Automated Image Annotation”, 2016; [http://arxiv.org/abs/1603.08486 arXiv:1603.08486].
- Tufts, Chris. “Gensim LDA: Tips and Tricks.” Mining the Details. N.p., 13 Aug. 2016. Web. 07 June 2017. [http://miningthedetails.com/blog/python/lda/GensimLDA/]
- Yulan He. 2016. Extracting topical phrases from clinical documents. In Proceedings of the Thirtieeth AAAI Conference on Artificial Intelligence (AAAI’16). AAAI Press 2957-2963.